Postdoctoral Research
In the Apostolou Lab, my research has focused on how epigentic regulators control gene expression during the earliest stages of mammalian development. Gene expression is controlled, in part, by cis-acting regulatory elements that act as landing sites for transcription factors that regulate the expression of nearby genes. In contrast to promoters, which are located proximal to the TSSs, enhancers often act over large genomic distances–often located 10s or even 100s of kilobases away from their target genes–and promote transcription by looping out the intervening DNA to come into close spatial proximity.
While many enhancers seem to exhibit exquisite specificity, a subset
form complex, nested interaction networks, which we call "hubs", in which dozens
of enhancers and promoters interact with one another in 3D space. Hubs seem to play an important
role in promoting high-levels of robust gene expression, and also allow for co-regulated gene
expression responses. Consequently, they play an critical role during cell fate transitions,
oncogenesis, immune response, as well as, really, any biological process that requires
robust, synchronized changes in gene expression (which is basically all of them).
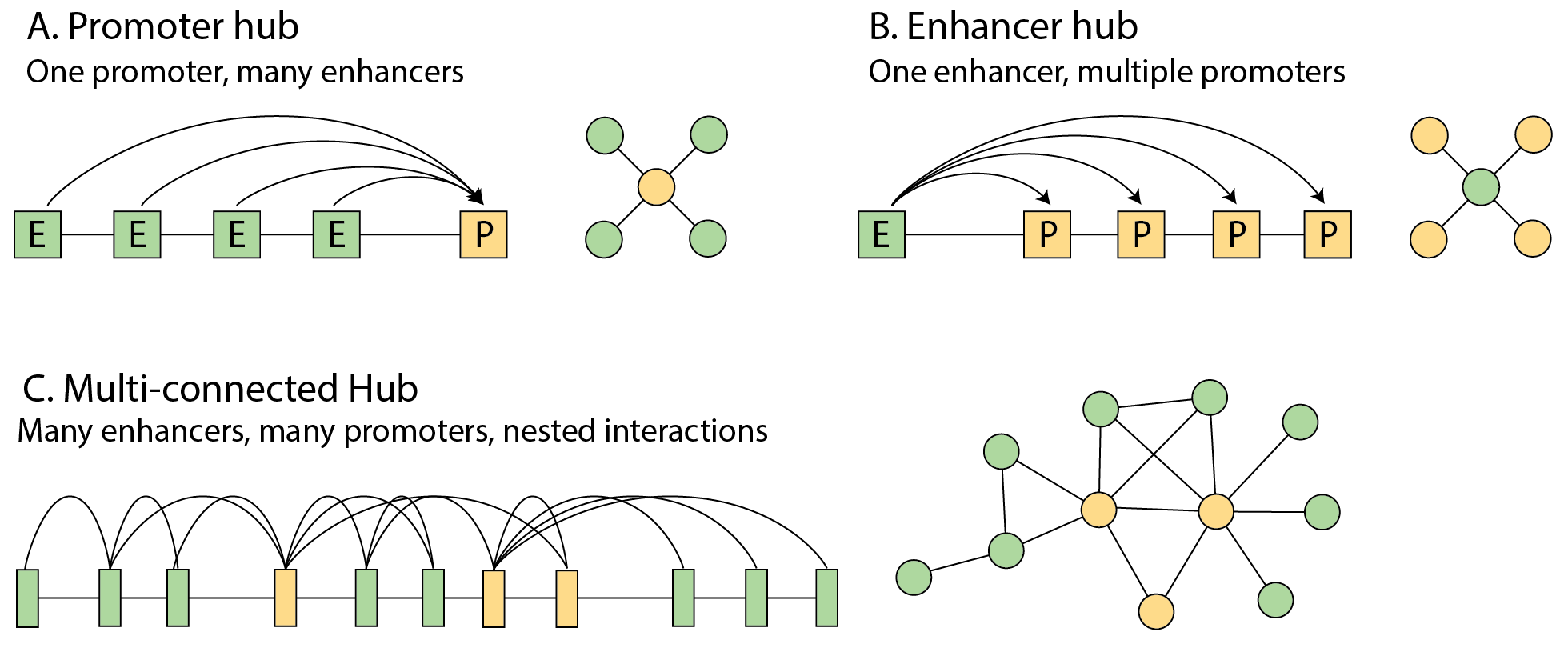
Hubs are organized in different ways, including A) one promoter
regulated by multiple enhancers and B) one enhancer acting on multiple
promoters. C) A subset of sites are organized into complex networks in
which multiple enhancers and promoters interact with one another in
complex, nested interaction networks ("hubs"). Multi-connected hubs
allow for co-regulated changes in gene expression during biological
processes which required coordinated changes in gene expression (ie most
of them) and can promote high-levels of gene expression. We reviewed
what's known about 3D hubs
here.
One of the projects that I've been involved with in collaboration with a postdoc in the Stadtfeld Lab. has focused on the the role of the histone methyltransferase EHMT2 in stabilizing cell identity in mouse embryonic stem cells. EHMT2 is a regulator of facultative heterochromatin through its ability to catalyze H3K9me2, a repressive histone mark, that is dynamic over the course of development. However, although studied for many years, the means by which EHMT2 acquires specificity during early embryonic development has remained poorly understood.
Using acute depletion of EHMT2, we found that loss of EHMT2 causes de-repression both of genes normally active later in development, which become preciously activated, as well as genes normally active earlier in development, which are normally shut off earlier in development. In other words, EHMT2 seems to stabilize the pluripotent state blocking both forward and reverse development. We further investigated what distinguished these two categories of genes. One of the more striking observations we made was that the genes the were normally active earlier in development, which are normally on during the onset of zygotic genome activation (ZGA) were organized into clusters of co-regulated loci. We call these "EHMT2-Coordinated Repressed Domains" (ECORDS).
ECORDS are distinguished from non-ECORDS both by their organzation into genomic clusters, as well by the way EHMT2 regulates them. To our surprise, we found that EHMT2 bound repetive LINE-1 sequences, which were not found enriched near non-ECORD genes. EHMT2-bound LINEs are interspersed throughout ECORDs and seem to act as nucleation sites that spread H3K9me2. LINEs had not previously been shown to act as silencers before. In fact, the specific classes we found to act as silencers had been recently reported to have the opposite role–acting as enhancers during ZGA. This suggests that there may be a molecular hand-off that occurs, in which LINEs begin by acting to promote ZGA, and then switch to silencers through differential binding of EHMT2. However, how this switch occurs, or whether it is gradual or sudden, is not yet known and an exciting area of future research.
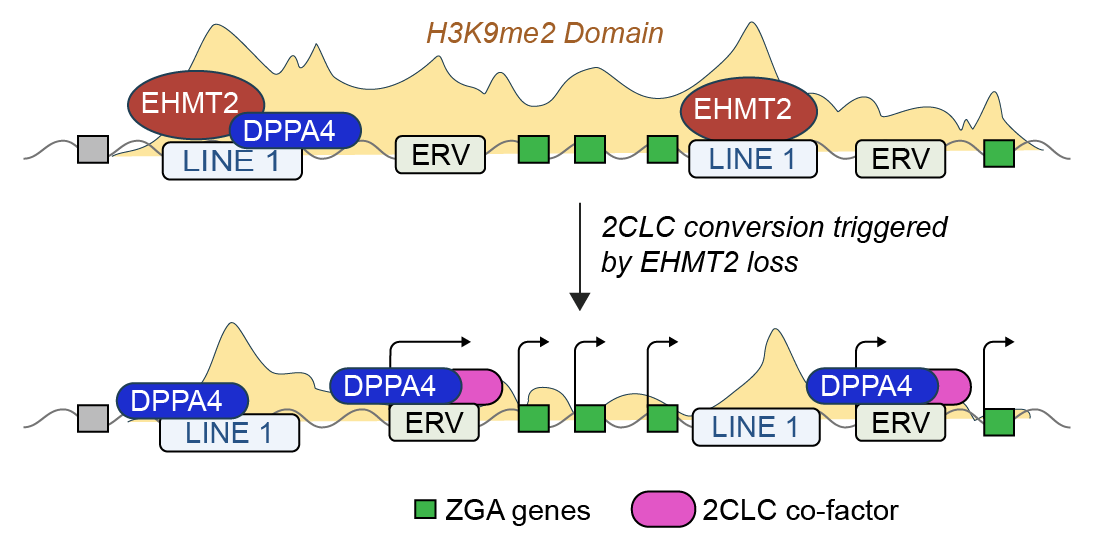
EHMT2 represses genes related to Zygotic Genome Activation (ZGA). In the absence of EHMT2, ESCs spontanously convert into "2-Cell Like Cells" (2CLCs), which resemble the earliest (totipotent) stage of mammalian development. The regulation of this state seems to occur through several class of repeatitive element. EHMT2-binding to LINEs nucleates broad H3K9me2 domains that spread from LINEs to cover the surrounding genomic loci. Spreading of H3K9me2 represses ZGA genes by inhibiting the activity of ERV repeats, which can as enhancers, as well, possibly, the promoters themselvers. However, The precise co-factors and transcription factors that mediate both the activation of ZGA genes in the absence of EHMT2, as well as the factors that recruit EHMT2 remain incompletely understood. You can read more about this in our recent paper.
Graduate Research
Overview
Broadly, my research in the McKay Lab focused on the epigenetic regulation of tissue- and temporal-specific gene expression profiles during development. My work used genomics to develop a high-level understanding of biological processes and combined it with targeted molecular biology experiments to validate and expand upon specific questions.
More specifically, I researched how ecdysone hormone signaling in the fruit fly, Drosophila melanogaster, elicits different metamorphic responses throughout the animal and over time. Canonically, it had been thought that pulses of ecdysone triggered gene expression cascades in tissues throughout the animal, acting to both synchronize and drive delopmental transitions forward when the animal was ready.
Molecularly, ecdysone achieves this effect by binding to its receptor, EcR, that, in turn, activates the expression of additional, primary response transcription factors. Canonically, EcR had always been positioned at the top of this hierarchy. My research showed that, EcR acted much more broadly than had previously been believed–it directly bound thousands of enhancers and promoters throughout the genome. By cloning and imaging the expression of patterns of its binding sites both over time, and during acute depletion of EcR, we we able to show that the binding was functional and regulated both the spatial and temporal activity of these enhancers.
Questions:
- How does the ecdysone receptor (EcR) coordinate temporal- and tissue- specific gene expression responses?
- Is its binding dynamic across time and between tissues?
- How does EcR regulate individual enhancers?
- Does it regulate their temporal activity?
- What dictates where EcR binds?

This enhancer from the Delta (Dl) locus is beautifully patterned. A) Browser shot of EcR CUT&RUN from Drosophila wings at the Delta locus. DlSOP is highlighted. B) DlSOP driving nls::GFP (green) compared to Ac, a marker of proneural clusters. C) DlSOP driving destabalized GFP (dsGFP) compared to Dl protein. When driving dsGFP, DlSOP is active only in a few cells that we believe correspond to the sensory organ precursor. You can see this figure, and other cool ones here.